Markus Strasser
Powered by 🌱Roam GardenBiological Expression Language (BEL)
BEL Biological Expression Language
BEL is a language for representing scientific findings in the life sciences in a computable form.
The BEL programming language is parse-able into a computable format that allows transformations and expansions for building a computable knowledge graph which can then be used in algorithms such as Reverse Causal Reasoning, Heat Diffusion or as prior knowledge for deep learning.
BEL is designed to represent scientific findings by capturing causal and correlative relationships in context, where context can include information about the biological and experimental system in which the relationships were observed, the supporting publications cited and the process of curation.
excerpt::
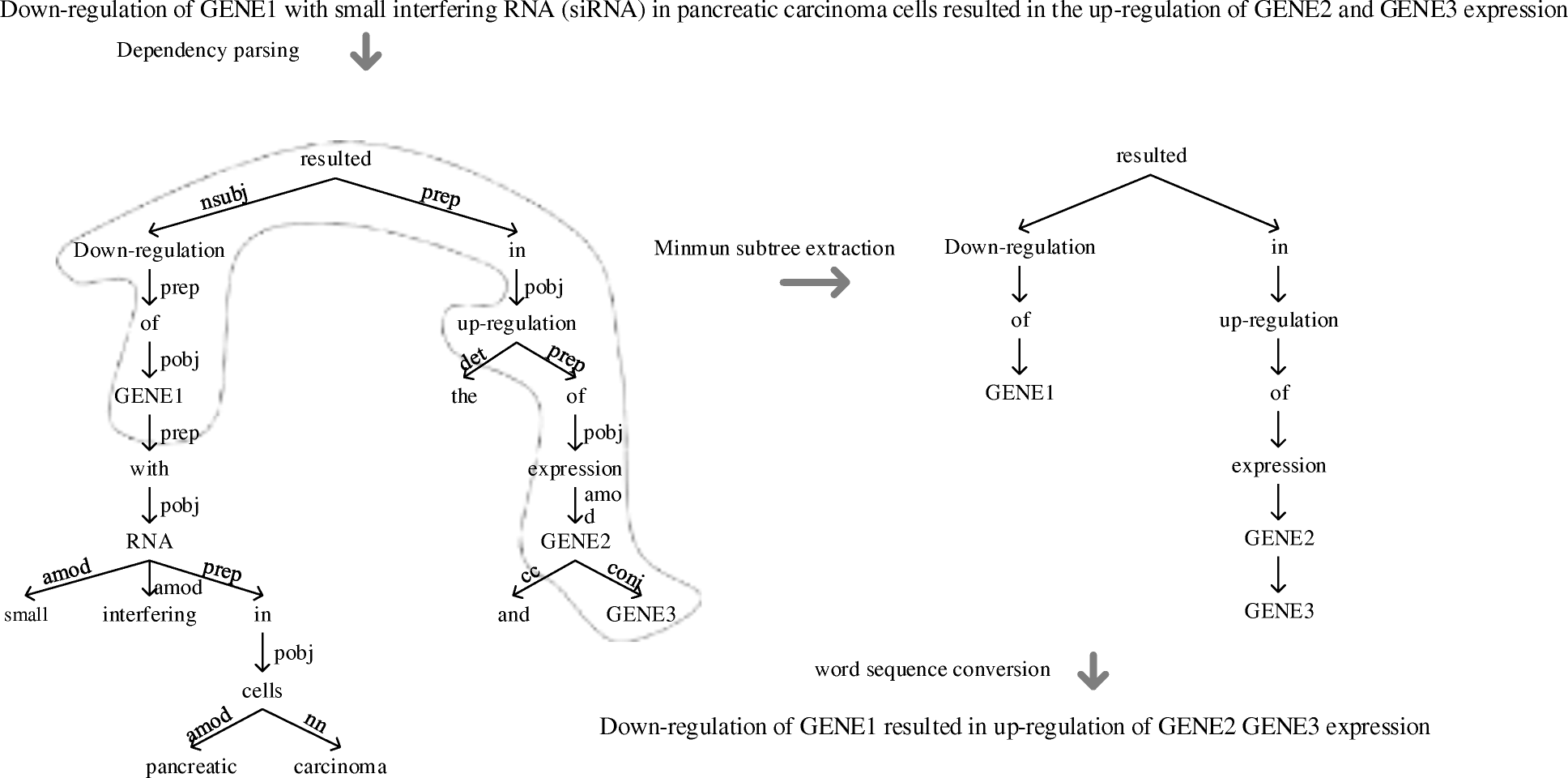
We first map the sentence-level BEL statements in the BC-V training corpus to the corresponding text segments, thus generating hierarchically tagged training instances.
A hierarchical sequence labeling model was afterwards induced from these training instances and applied to the test sentences in order to construct the BEL statements.
F-measure improvement of 31.6%
Unlike the previous relation extraction task, where a relationship is purely between two entities, the BEL task aims to discover the hierarchical relations between biomedical entities, meaning that the relationship (increases or decreases) can hold among multiple entities and complex biomedical functions (such as complex() or tloc()) can also be involved.
Taking the following sentence together its corresponding BEL statement extracted from BC-V corpus as an example, it illustrates a hierarchical relation involving three entities, one function and one relationship: the catalysis of the protein IL-2 increases the complex between the protein LYN and the protein IL2RB.
The association of __lyn__ with __IL-2Rbeta__ was markedly elevated by __IL-2__ stimulation. (PMID: 11131153).
cat(p(HGNC:__IL2__)) increases complex(p(HGNC:__LYN__), p(HGNC:__IL2RB__)).
BEL Example
“Shear stress enhances expression of the gene encoding the endothelial nitric oxide synthase (eNOS) and further stimulates its enzymatic activity, leading to physiologic low concentrations of nitric oxide (NO) within endothelial cells [44–46]. This continuous generation of NO prevents the apoptosis of endothelial cells, thereby protecting the endothelial monolayer from injury [47,48].”
Is converted into BEL Assertions:
biologicalProcess(GO:“response to fluid shear stress”) increases rnaAbundance(HGNC:NOS3)
biologicalProcess(GO:“response to fluid shear stress”) increases activity(proteinAbundance(HGNC:NOS3), molecularActivity(cat))
activity(proteinAbundance(HGNC:NOS3), molecularActivity(cat)) directlyIncreases abundance(CHEBI:“nitric oxide”)
abundance(CHEBI:“nitric oxide”) decreases biologicalProcess(GO:“apoptotic process”)
the abbreviated BEL Assertions are:
bp(GO:“response to fluid shear stress”) -> r(HGNC:NOS3)
bp(GO:“response to fluid shear stress”) -> act(p(HGNC:NOS3), ma(cat))
act(p(HGNC:NOS3), ma(cat)) => a(CHEBI:“nitric oxide”)
a(CHEBI:“nitric oxide”) -| bp(GO:“apoptotic process”)